-
-
Access Free Student Software
Ansys empowers the next generation of engineers
Students get free access to world-class simulation software.
-
Connect with Ansys Now!
Design your future
Connect with Ansys to explore how simulation can power your next breakthrough.
Countries & Regions
Free Trials
Products & Services
Learn
About
Back
Products & Services
Back
Learn
Ansys empowers the next generation of engineers
Students get free access to world-class simulation software.
Back
About
Design your future
Connect with Ansys to explore how simulation can power your next breakthrough.
Free Trials
ANSYS BLOG
April 15, 2024
Validating Your Gut Feelings With Simulation
Consider the stomach. It doesn’t garner the respect that the brain does. It also ranks lower than the heart on every Internet pundit’s top 10 list of important internal organs. Yet everyone remembers the assertion, even if they don’t remember who said it, that an army marches on its stomach. The line is clever only because of the complicated truths it contains: The stomach is not as glamorous as the brain or the heart, but when it’s full we’re happy and when it’s empty, we’re not. We put a wide variety of things into it — liquids and solids, foods and medicines — and generally it goes about its business of breaking things down and facilitating the absorption of nutrients and medications. At the same time, it can be upset easily if we put things into it that disagree with it.
All of this we know from experience, but past experience is not always predictive. Around the world, food companies are striving to develop new foods that can sustain a hungry world more effectively and at a lower cost. Pharmaceutical companies are striving to develop new medications that can cure or lessen the effects of diseases. The success of innovations in these fields will often be determined by what happens in the stomach.
The difficulty, though, lies in developing an understanding of what will actually happen when a novel food or medication arrives in the stomach.
“In vivo studies of digestion are very complex, expensive, and time consuming,” notes Dr. Xinying Liu, a research associate in the School of Chemical and Biomolecular Engineering of the University of Sydney. “They are invasive and very difficult to perform, requiring both specific technical skills and the prior approval of an ethics and human subjects review board.”
In vitro models of the digestive system can address some of these issues, but the complexities of the stomach — with its ability to layer foods for optimal digestion, its moving walls, and its complex acid and enzyme interactions — are quite difficult to reproduce accurately in an in vitro model.
For all these reasons, Dr. Liu and Dr. David F. Fletcher, an adjunct professor at the School of Chemical and Biomolecular Engineering at the University of Sydney, have been working to refine a computer model, also known as an in silico model, of the digestive system. With an accurate, operational in silico model of the digestive system and all its component parts — from the esophagus through the stomach and into the small and large intestine — food and drug developers, as well as other researchers in related commercial and academic fields, could gain insights to how the digestive system might respond to new or modified foods and drugs long before they actually engage human subjects to test them.
Mixing behavior within the stomach with initial fluid volume divided into five bands.
Acid distribution within the stomach with acid secreted from the stomach wall.
The simulations above used the smooth-particle hydrodynamics (SPH) model of the the Commonwealth Scientific and Industrial Research Organization (CSIRO).
Developing and Validating an In Silico Model of the Digestive System
Liu and Fletcher did not start from nothing when developing their simulations of the digestive system. Some work had already been done within Australia’s government-funded research entity, the Commonwealth Scientific and Industrial Research Organization (CSIRO) by Drs. Simon Harrison and Paul Cleary. The efforts at CSIRO had involved the development of a stomach simulation using tools based on a smooth-particle hydrodynamics (SPH) approach. While the SPH-based approach had its strengths — particularly for simulating a highly dynamic system like a stomach — Liu and Fletcher wanted to verify and validate these simulations using a different set of approaches. These efforts became the core of Liu’s doctoral dissertation, which was shepherded to completion under Dr. Fletcher’s supervision.
Using a range of tools from Ansys — including Ansys Mechanical, Ansys Fluent, and Ansys System Coupling — Liu and Fletcher set about validating and, as necessary, refining the SPH-based models. One of the first simulations they developed involved simulating peristaltic flow over time. This is crucial to any digestive model because peristalsis effectively mixes food and nutrients and pushes the contents of the stomach through the digestive tract. Their simulation involved modeling the contraction and expansion of muscle fibers within the digestive tract to create a wave-like motion that influences the contents of the digestive tract. As the peristaltic wave propagates, it mixes the materials within the digestive tract and moves them along. At the same time, the physical characteristics of the stomach, the pylorus valve, and different sections of the intestines introduce variables such as velocity, pressure, and material concentrations — all of which must be accommodated in any complete simulation of the digestive process.
Liu and Fletcher used the finite volume method (FVM) embedded in Fluent and the SPH code the team at CSIRO had built, and then compared the volumetric flow results of both simulations to results from an analytic solution. This work exposed issues with the SPH code that resulted in tensile instabilities. Using the results gained through FVM, the team showed that by adding a pressure offset component in the SPH code, the tensile instability could be resolved.
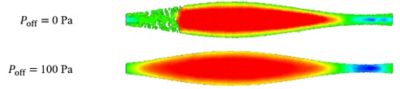
Without incorporating a pressure offset into the smooth-particle hydrodynamics (SPH) code, as illustrated in the lower simulation, the fluids in the top simulation appear to cavitate, which does not occur in reality.
Modeling the Dynamics of the Stomach
Building on their experience with modeling peristaltic flow, Liu and Fletcher went on to verify the findings of the SPH-based model of the stomach itself, with specific attention to the manner in which the stomach mixes and separates different types of solids and liquids. Because the acids and enzymes in the stomach break down different types of food and medicine into smaller particles that can be digested more completely in the intestines, different types of food have different residence times in the stomach which can be achieved via layering, so that that the acids and enzymes within the stomach have time to break them down.
From a simulation standpoint, any model of digestion within the stomach must accurately reflect the mechanisms within the stomach that orchestrate this layering. Solids will end up on the lowest level of the stomach; liquids will sit on top of solids; fats will sit on top of liquids and provide a buoyant layer that drug developers can use to float medications that require prolonged exposure to the gastric conditions of the stomach.
Simulating the physics of the stomach that create and maintain this layering is complicated. Moreover, incorporating the effects of stomach acids and enzymes that break down the components of the different layers (which will break down at different rates in different layers) adds further complication to any modeling efforts. The models that Liu, Fletcher, and the CSIRO team analyzed were focused on the physics of the buoyancy-driven separation of these layers.
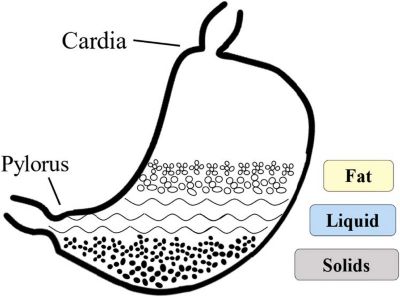
Solids end up on the lowest level of the stomach; liquids sit on top of solids; fats sit on top of liquids.
They did not yet attempt to model the complexities of real-world food digestion (with different types of food and numerous digestive layers) but focused, rather, on simulating the manner in which just two liquid layers — one fatty and one aqueous — would separate under gravity as a consequence of what is called Rayleigh-Taylor instability, which arises from the differences in density and the surface tension that naturally try to stabilize the layers. In the simulation, the aqueous layer begins above the fatty layer (with liquid oil representing a fatty layer) and the Rayleigh-Taylor instabilities naturally initiate a buoyancy-driven flow that causes the layers to invert so that the fatty layer ends up above the aqueous layer.
As with the peristalsis simulations, Liu and Fletcher relied on the FVM methodology of Fluent to replicate and validate the SPH model. In the SPH model, the oil and water invert over the course of six seconds and the details of the Rayleigh-Taylor instabilities that initiate this inversion are quite distinct.
In the smooth-particle hydrodynamics (SPH) simulation, the oil and water layers invert over the course of six seconds and the details of the Rayleigh-Taylor instabilities are quite visible. However, the smooth surface between the oil and water layers that should exist at the completion of this inversion does not appear in the SPH simulation.
In the finite-volume method (FVM) simulation, the Rayleigh-Talor instabilities are less well-defined but the oil and aqueous layer boundary is far more distinct at the end of the simulation.
The FVM simulation conducted using Fluent verifies many aspects of the SPH model but also highlights distinct differences. In the FVM simulation, the Rayleigh-Talor instabilities are less well-defined, but the oil and aqueous layer boundary is more accurately presented at the conclusion of the simulation. Repeated running of the SPH model — even for longer durations — never results in as distinct a layer between the newly inverted liquids.
“The smoothed buoyancy force may become less effective in the SPH model when the thickness of the fluid fragments is smaller than the kernel width,” notes Liu. “It may be that the density difference is smoothed over the kernel diameter, resulting in a smaller magnitude which results in a reduced buoyancy force — and the smaller the region of fluid, the worse this effect becomes. As for the lack of detail in the growth of the Rayleigh-Taylor instabilities in the FVM model, that may be the result of errors induced by the grid that was used rather than any specific shortcoming in the FVM approach itself.”
Marching Orders
Together with their CSIRO colleagues, Liu and Fletcher have gone on to develop SPH simulations of peristaltic movement in the stomach wall and models mimicking the mixing and distribution of acid within the stomach; they have also developed simulations modeling the effect of viscosity on the speed with which the fluid layers within the stomach are mixed. Further models must still be built and validated before a complete and accurate in silico model of the digestive system can be said to exist, but the models developed so far appear to be on the right track.
“It is quite tricky to get set up a good simulation of the stomach,” says Dr. Fletcher. “You need the tools that can model non-Newtonian flow — with particles, complex rheology, complex chemistry, moving walls, free surfaces, and the like. There is a whole range of physics that is needed, too, and that is where the ability to bring together mechanical and fluids — and then the ability to model particles within fluids — becomes critical. We need to be able to model everything from very small particles that move with the fluids all the way up to large chunks that break down over time and as a function of the physics and the chemistry of the gastric environment. Ansys has the technology to do all this with its wide range of solvers, including SPH, so we can see where this is going and what the future will hold.”
These first encouraging results show that it is possible to reliably model the complexity of the digestive system. It paves the way toward future digital twins of the entire digestive system, a necessary step to optimize the absorption of drugs throughout the stomachal wall, test the consumption of new nutrients ... and allow us to keep enjoying food.
Learn how Ansys Mechanical, Ansys Fluent, and Ansys System Coupling can help you with your healthcare innovations.